Contributions
The following list includes only slides and tutorials where the individual has been added to the contributor list. This may not include the sum total of their contributions to the training materials (e.g. GTN css or design, tutorial datasets, workflow development, etc.) unless described by a news post.
GitHub Activity
Tutorials
- Transcriptomics / Bulk RNA Deconvolution with MuSiC
- Transcriptomics / Downstream Single-cell RNA analysis with RaceID
- Transcriptomics / Analysis of plant scRNA-Seq Data with Scanpy
- Transcriptomics / Pre-processing of Single-Cell RNA Data
- Transcriptomics / Understanding Barcodes
- Transcriptomics / Clustering 3K PBMCs with Scanpy
- Transcriptomics / Pre-processing of 10X Single-Cell RNA Datasets
- Transcriptomics / Trajectory Analysis using Python (Jupyter Notebook) in Galaxy
Slides
- Transcriptomics / An introduction to scRNA-seq data analysis
- Transcriptomics / Dealing with Cross-Contamination in Fixed Barcode Protocols
- Transcriptomics / Clustering 3K PBMCs with Scanpy
- Transcriptomics / Plates, Batches, and Barcodes
- Transcriptomics / Introducción al análisis de datos de scRNA-seq
- Transcriptomics / Una introducción al análisis de datos scRNA-seq
News
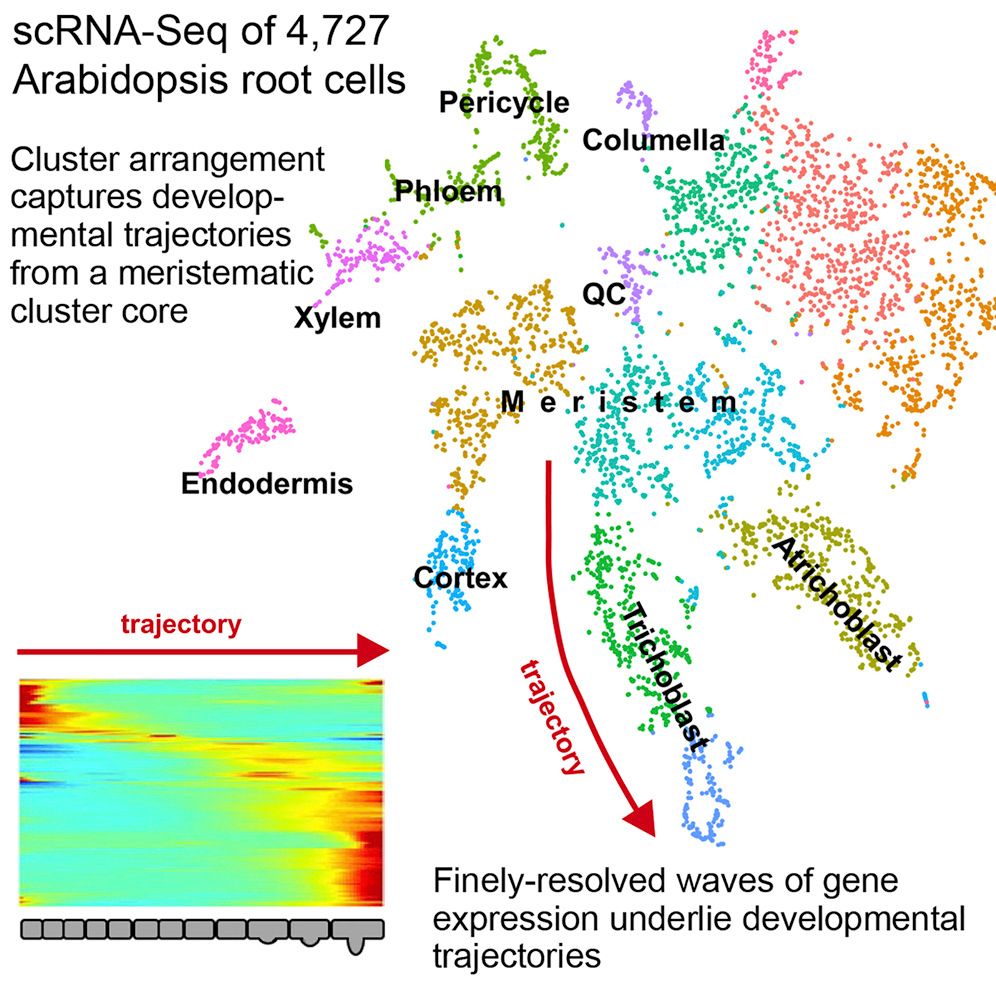
Authors:
Single cell RNA-seq analysis is a cornerstone of developmental research and provides a great level of detail in understanding the underlying dynamic processes within tissues. In the context of plants, this highlights some of the key differentiation pathways that root cells undergo.
Full Story